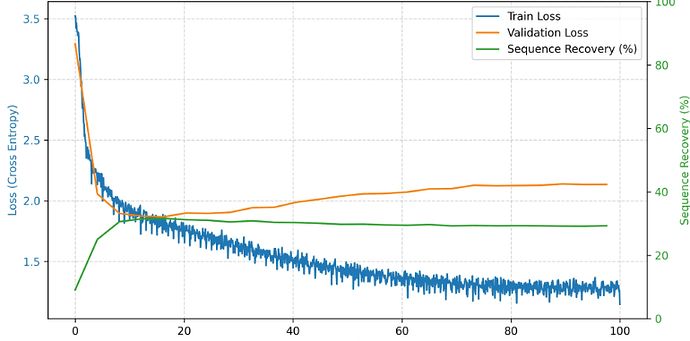
Hey thank you very much for your answer. I am trying to go through the most important points. I also tried to print some examples during training to check the padding. My actual usecase is that I am translating tokenized protein structures to sequences, so I am not using text or sentences (The model that I am using is pretrained on this protein data as well). E.g. here I print out the Tokens that the model gets, the models prediction, the label/gold and the padding and attention masks. A problem might also be that some sequences are very uninformative and have lots of repeating tokens, so I am now experimenting with filtering the data.Here are some examples:
--- save-pred example batch0-idx0 ---
TOKENS : ['d', 'v', 'a', 'v', 'q', 'a', 'v', 'v', 'v', 'v', 'y', 'v', 'y', 'y', 'v', 'v', 'v', 'v', 'v', 'q', 'v', 'v', 'q', 'c', 'v', 'l', 'l', 'l', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'c', 'y']
PRED : ['4', '12', '2', '2', '4', '4', '4', '4', '12', '2', '5', '4', '4', '13', '17', '4', '17', '4', '12', '7', 'LABEL_0', '12', '5', '7', '5', '5', '5', '7', 'LABEL_0', '12', '12', '12', '5', '3', '5', '5', '5', '12', '5', '12']
GOLD : ['10', '8', '2', '18', '13', '9', '12', '15', '-100', '15', '5', '12', '19', '3', '9', '14', '7', '3', '17', '13', '12', '8', '15', '-100', '-100', '14', 'LABEL_0', '14', '19', '3', '16', '3', '5', '15', '14', '5', 'LABEL_0', '17', '8', 'LABEL_0']
PAD POS: [178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
ATT=0 : [178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
SEQ LEN: 211, VALID: 178
--- save-pred example batch0-idx1 ---
TOKENS : ['d', 'v', 'a', 'v', 'q', 'a', 'v', 'v', 'v', 'v', 'y', 'v', 'y', 'y', 'v', 'v', 'v', 'v', 'v', 'q', 'v', 'v', 'q', 'c', 'v', 'l', 'l', 'l', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'c', 'y']
PRED : ['8', '17', '9', '17', '17', '5', '5', '17', '17', '17', 'LABEL_1', '17', '17', '2', '2', '5', '5', '17', '17', '17', '17', '5', '16', '17', '19', '13', '9', '7', '8', '8', '12', '8', '5', '17', '17', '5', '3', '4', '5', '17']
GOLD : ['10', '19', '5', '11', '18', '5', '14', '4', '7', '14', '17', '11', '9', '15', '16', '5', '2', '7', '8', '17', '3', '3', '19', '2', '3', '3', '9', 'LABEL_0', '8', '8', '18', '9', '5', '15', '14', '5', '9', 'LABEL_0', '7', '19']
PAD POS: None
ATT=0 : None
SEQ LEN: 211, VALID: 211
--- save-pred example batch0-idx2 ---
TOKENS : ['d', 'v', 'a', 'v', 'q', 'a', 'v', 'v', 'v', 'v', 'y', 'v', 'y', 'y', 'v', 'v', 'v', 'v', 'v', 'q', 'v', 'v', 'q', 'c', 'v', 'l', 'l', 'l', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'c', 'y']
PRED : ['15', '17', '4', '5', '13', '12', 'LABEL_0', '5', '13', '17', '7', '9', '5', '2', '15', '11', '15', '5', '2', '2', '9', '9', '17', '17', '5', '11', 'LABEL_0', '17', '3', '3', 'LABEL_0', '5', '2', '2', '17', '13', '2', '16', '5', '13']
GOLD : ['14', '4', '3', '16', '3', '3', '16', '14', '8', '9', '3', '7', '15', '5', '10', '12', '9', '5', '2', '9', '4', '-100', '7', '14', '4', 'LABEL_0', 'LABEL_0', '14', '15', '12', '3', '4', '8', '8', '15', '12', 'LABEL_0', '17', '14', '8']
PAD POS: [169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
ATT=0 : [169, 170, 171, 172, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188, 189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200, 201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
SEQ LEN: 211, VALID: 169
--- save-pred example batch0-idx3 ---
TOKENS : ['d', 'v', 'a', 'v', 'q', 'a', 'v', 'v', 'v', 'v', 'y', 'v', 'y', 'y', 'v', 'v', 'v', 'v', 'v', 'q', 'v', 'v', 'q', 'c', 'v', 'l', 'l', 'l', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'v', 'c', 'y']
PRED : ['12', '8', '5', '13', '18', '7', '7', '13', '12', '5', '12', '3', '3', '9', '14', '7', '7', '8', '14', '17', '9', '14', '19', '19', '14', '19', '9', '5', '8', '2', '17', '2', '8', '2', '8', '8', '8', '15', '5', '8']
GOLD : ['14', 'LABEL_0', '8', '17', 'LABEL_0', '10', '15', '-100', '4', '3', '12', '-100', '3', '19', '7', '14', '19', '2', '9', '9', '3', '8', '11', '7', '2', '7', '17', '14', '8', '14', '9', '11', '14', '12', '9', '16', '9', '15', '3', '8']
PAD POS: [201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
ATT=0 : [201, 202, 203, 204, 205, 206, 207, 208, 209, 210]
SEQ LEN: 211, VALID: 201